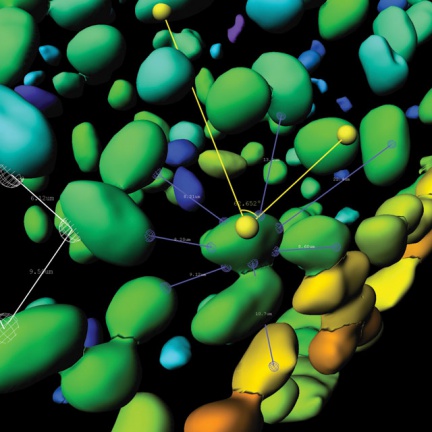
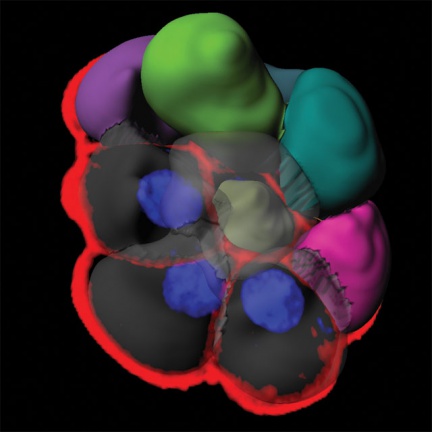
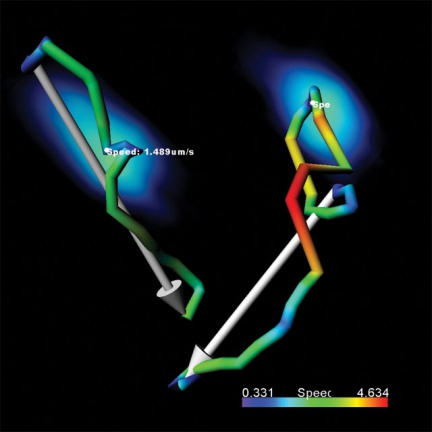
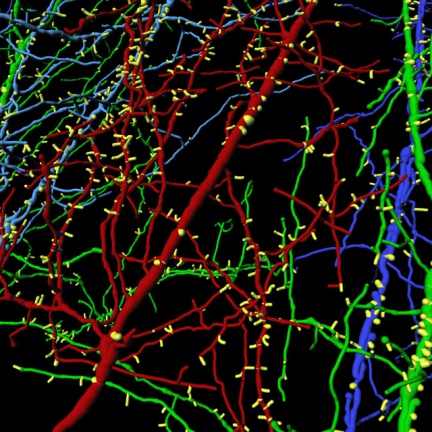
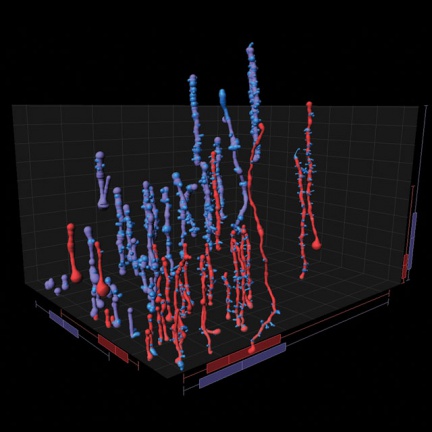
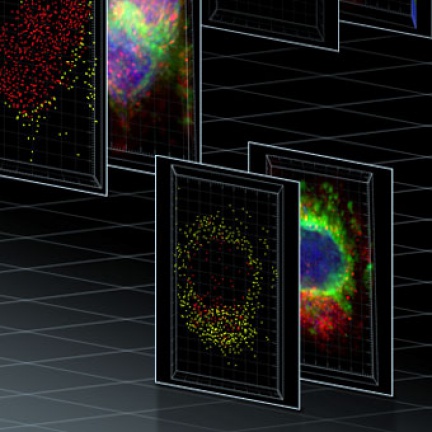
Imaris software can work with most of the data formats used.
| Imaris file format |
*.ims |
| Compatibility (file format) |
*.tif, *.tiff, *.xml, *.h5, *.r3d, *.dv, *.ipl, *.kinetic, *.cxd, *.lif, *.stk, *.ics, *.nd2, *.oib, *.vsi, *.zvi, *.czi, and many more |
| Operating system |
MS Windows, MAC OS, MAC OS X |
Accessories
For image data analysis, Imaris includes many special modules that are tailored to specific applications. All available analysis tools are represented by a simple wizard that contains multiple steps for specifying the required task.
Deconvolution module
The ClearView GPU Deconvolution module is used for deconvolution - deconvolution algorithms for various microscopy methods (spinning disk confocal microscopy, TIRF, widefield fluorescence, brightfield, laser scanning confocal microscopy) are integrated directly into the Imaris software . The supported GPU processing modes are CUDA and OpenCL. A graphics card driver update is required to run deconvolution. NVIDIA cards with drivers 411.31 and later use CUDA for processing, all AMD cards and NVIDIA cards with older drivers use OpenCL. Macs running MAC OS 10.14 will only use OpenCL, despite the presence of an NVIDIA card, as there is not yet a supported driver for this graphics card for MAC OS 10.14. Older workstations or graphics cards may not support deconvolution at all. CPU Deconvolution can be used instead.
A list of tested GPUs and more detailed information on recommended workstation configurations for working with Imaris software and running deconvolution analysis can be found here.
The following is a complete list of available modules for Imaris software:
"Analyzes extremely large and complex microscopic images quickly and easily"
A basic image analysis tool that is part of the Imaris core and adds the ability to measure object geometry and intensity to the software.
- Interactive and fast imaging of huge surfaces and millions of point objects
- Analysis of surfaces and point objects within voluminous datasets
- Classification and labeling of surfaces and point objects using Machine Learning classifier or interactive filters
- Report and compare parameters based on detected classes
- Shape and intensity measurements based on individual channels in 3D/4D
- Statistical data
- Calculation of distance and overlap between objects
- Measurement of attraction and repulsion of detected objects compared to random distribution
- Creation and measurement of 3D objects based on 2D contours
ImarisCell
"Explains relationships between cells"
Comprehensive analysis of groups of cells and individual cells and their components on a cellular basis. Analysis of image data capturing cell cultures, providing statistical outputs categorized by basic cell morphology.
- Analysis of relationships between cells and their organelles
- Measurement of mechanical and structural cellular functions (cell-to-cell communication)
- Detection of cells based on cytoplasmic or plasma membrane staining
- Use of intuitive units based on cell morphology (nuclei, vesicles, membranes)
- Detection and classification of multiple populations of vesicular objects
- Study of cell behaviour (in 2D/3D/4D)
- Intuitive, structured, advanced step-by-step analysis guide
ImarisColoc
"Isolates, visualizes and quantifies colocalized regions"
Colocalization Analysis. A basic tool for determining the touching areas and overlapping points of signals from different color channels(colocalization).
- Colocalization rate
- Selection of different colocalization methods (including automatic mode based on established algorithms)
- Real-time statistical data acquisition
- Data after colocalization presented as a new color channel (3D or 4D)
- Option to expand or narrow the calculated histogram area
- Perform colocalization analysis on specific ROIs
- Colocalization of the entire time series in fewer steps
ImarisTrackLineage
"Tracks object movement and detects cell division"
Object motion analysis. Mapping cell differentiation. A great tool for 3D and 4D object tracking and time-lapse image processing capturing the progression of biological processes(live cell imaging) from a single original cell(progenitor cell) to the final stages of cell division.
- Automatic object tracking in 2D, 3D + time
- Automatic detection of cell division
- Choice of several specific tracking algorithms for different types of movement
- Fast processing of thousands of objects at a single time point, fast processing of thousands of time points
- Interactive creation and editing of tracks and tracked objects
- Analysis of speed, shape, intensity, size and other parameters
- Determine cell cycle time and generation, visualize the lineage of cell populations
- Automatic correction of translational and rotational drift using the Reference Frame function
- Analysis of very long time experiments - time lapse
FilamentTracer
"Visualizes and analyzes neurons and other filamentary structures in 3D"
Detection, 3D visualization, tracking and analysis of filaments. Analytical tool mainly focused on neurobiological data, with the possibility to apply algorithms to different filamentary structures.
- Detection and tracking of filamentary structures (dendrites, cytoskeleton, tubules,...)
- Algorithms for automatic analysis and interactive 3D tracking methods (Wizard Guided Automatic or AutoPath and AutoDepth methods)
- Automatic detection and determination of morphological characteristics of dendritic spines
- Facilitating tracing in dense neural networks using Imaris Torch
- Determination of statistical data - branch length, diameter, area, volume, dendritic spine density, fiber topology, etc.
- Direct interaction with the entire filament, individual branches, segments or specific points
- Possibility of 3D visualization of the filamentary structure and dendritic spines (resolution e.g. colour, size) together with other, non-filamentary objects
- Tracking and detection of shape and position changes over time (in combination with Imaris TrackLineage)
- Possibility of manual processing or manual final editing
ImarisVantage
"Built for interpreting scientific discoveries"
Option for sophisticated presentation of acquired data and interpretation of results using interactive multidimensional plots comparing optional statistics.
- Graphical display of data - 1D/2D/3D/4D (1 parameter: side-by-side plot, point diagrams for 2 parameters, object gallery view & scatterplots, Box and Whisker Plots, 5-Number summary)
- Comparison of two or more groups of data (control and test groups), comparison between classes
- Use calculated parameters to determine object dimensions, color coding and scaling
- Obtaining Wilcoxon, T-test, F-test and Kolmogorov-Smirnov results and exporting these results for further statistical analysis
- Statistical calculations, trend and outlier detection
- Creation of spatial object interaction graphs and event time diagrams
ImarisBatch
"The ultimate Imaris productivity tool"
Simultaneous data processing and analysis ("in one batch"). Time saving in image processing and analysis by automatically applying a defined protocol to a large group of 2D/3D+ time frames.
- Reproduction of accurate analysis procedures
- Interactive creation of a universal protocol for bulk analysis of "n" images
- Application of the protocol to different image datasets simultaneously
- Seamless integration into Imaris workflow including Machine Learning classification
- Unified image processing pipeline
- Possibility to compare statistical data in the Vantage module
ImarisXT
"Expands the horizons and possibilities of analysis in Imaris"
User extension of image analysis capabilities using common programming languages. ImarisXT is an application programming interface that allows you to add custom functions to Imaris and exchange data between Imaris and Matlab/Java/Python in two ways.
- Customizing analysis in Imaris
- Ability to add custom functions to Imaris
- Extension of basic Imaris functionsby adding a user's own plug-in
- Compatibility with Matlab/Java/Python/ImageJ/FIJI
- Public web platform for sharing user extensions - Imaris Open
- Extension capabilities for a large number of applications including super-resolution, cell tracking, filament tracing, GPU object detection deconvolution, batch analysis, collocation, etc.
- Development of custom algorithms for image processing and segmentation
- Created by members of the "ImarisXT Developer Program"
- Free download of more than 70 XTenstions